Navigation
What is gutMEGA
The gutMEGA is a human gut metagenome database, which collated 182 literatures for human gut microbiome research. In total, 59,132 quantification events for 6,457 taxonomies at 7 different levels (kingdom, phylum, class, order, family, genus, and species) under 776 conditions were collected and integrated in the database, and the details about the samples and experiments were also curated. Various browse and search options were provided in the database, and the detailed information was organized and visualized for queries.
Metagenome means the sum of all the genetic materials in a certain environment. Human gut metagenomics mainly study the intestinal microbiota in human under different conditions and reveal its role in diseases, tumors, growth and development, mental health, etc.
How to use gutMEGA
Quick Search
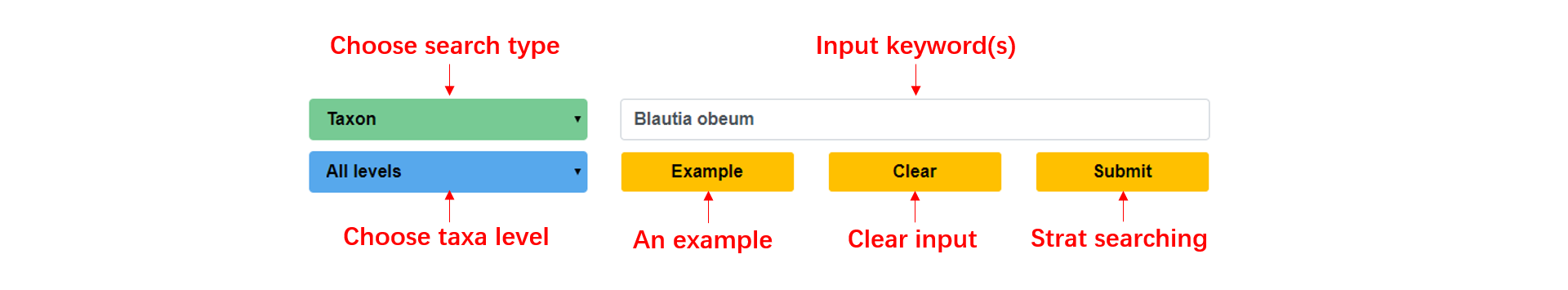
In the home page of gutMEGA, users can start a quick search with the keyword(s) of interest or simply click the search button to see the search results of an example for which we provided in the input field.
Browse
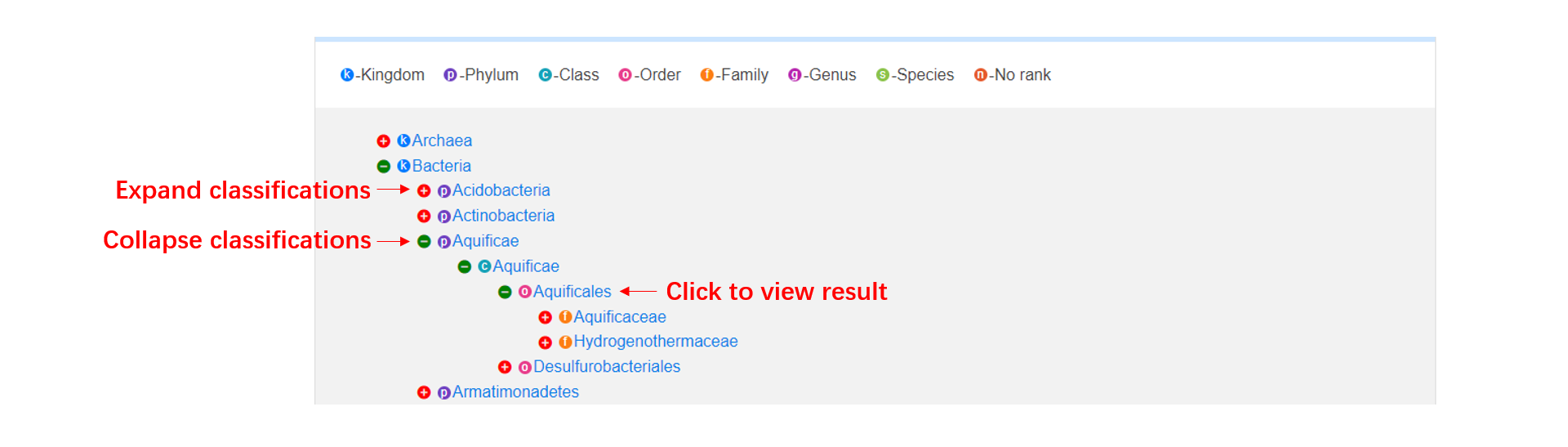
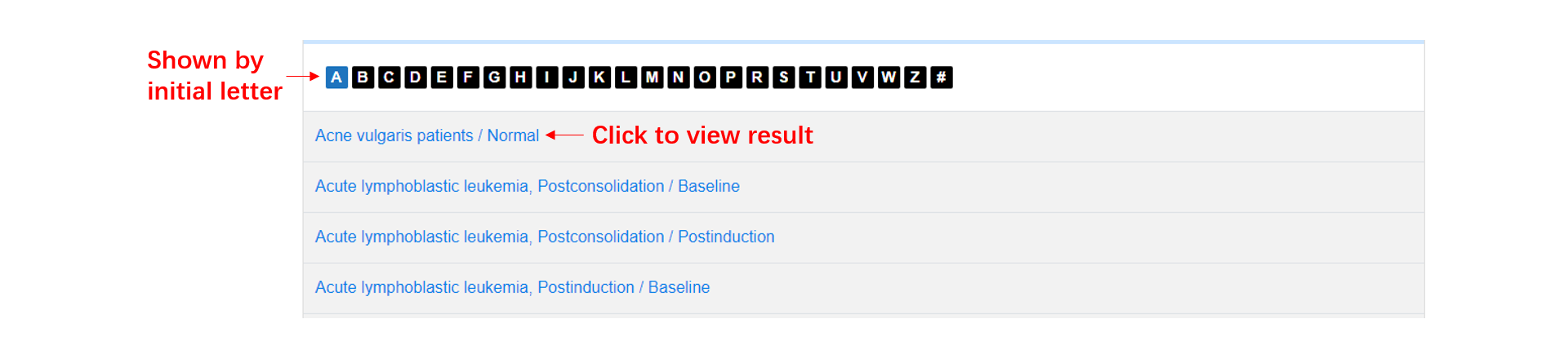
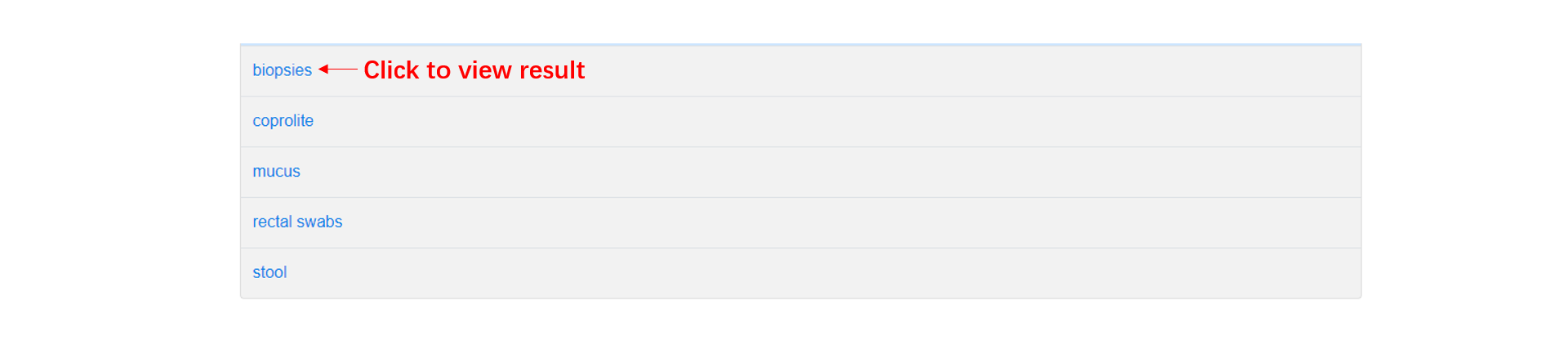
The browse page consists of three parts: (1). In taxonomy table, users can click '+' button to view the detailed classifications, and browse the results by clicking the taxon. (2). In condition table, we show all the conditions by their initials, users can click one condition to browse the results. (3). In sample table, users can click one sample to browse the results.
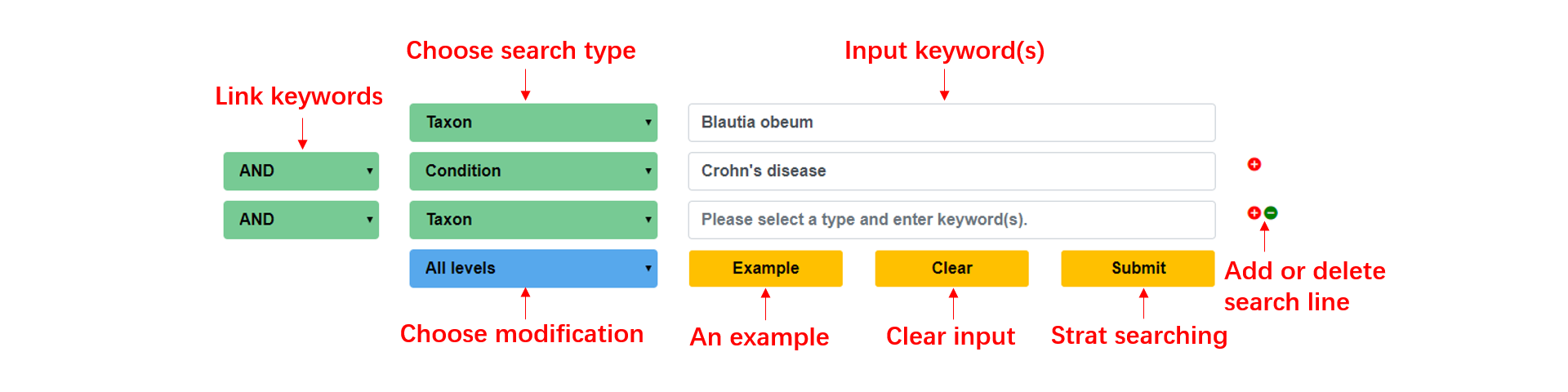
Advanced Search
In the search page, we provide an advanced search capability, users can add more search modules by '+' button, and delete redundant search modules using the '-' button, and choose 'AND', 'OR' or 'NOT' to connect the keywords for a more accurate multiple search, also, we provide an example button for users as a search paradigm.
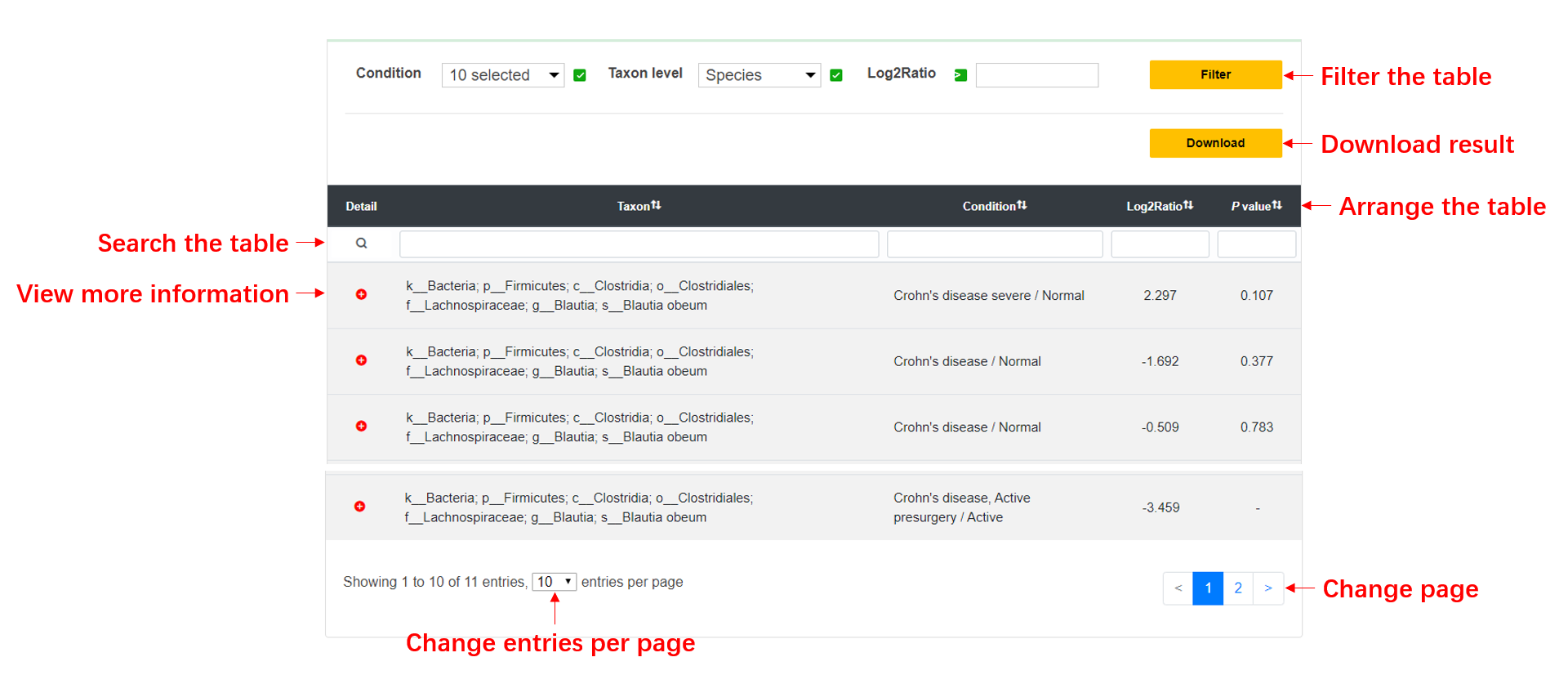
Result
In the Result page, users can view the search or browse results, and further filter them by the options we provide. More information about experiment is also displayed while users click the 'Detail' button.
Metagenome reference database
Greengenes
A 16S rRNA gene database addresses limitations of public repositories by providing chimera screening, standard alignment, and taxonomic classification using multiple published taxonomies.
http://greengenes.lbl.govSILVA
SILVA provides comprehensive, quality checked and regularly updated datasets of aligned small (16S/18S, SSU) and large subunit (23S/28S, LSU) ribosomal RNA (rRNA) sequences for all three domains of life (Bacteria, Archaea and Eukarya).
https://www.arb-silva.deRDP
RDP provides quality-controlled, aligned and annotated Bacterial and Archaeal 16S rRNA sequences, and Fungal 28S rRNA sequences, and a suite of analysis tools to the scientific community.
http://rdp.cme.msu.eduNCBI
The Taxonomy Database is a curated classification and nomenclature for all of the organisms in the public sequence databases. This currently represents about 10% of the described species of life on the planet.
https://www.ncbi.nlm.nih.gov/taxonomy